Formulation and Delivery - Biomolecular
Category: Poster Abstract
(M1330-11-75) Reversible, Covalent DNA Condensation Approach via Chemical Linkers for Enhanced Gene Delivery to Nucleus
Monday, October 23, 2023
1:30 PM - 2:30 PM ET
- AT
Aaron Tasset, BS
University of Texas at Austin
Austin, Texas, United States - WW
Wenliang Wang, Ph.D.
University of Texas at Austin
Austin, Texas, United States - IP
Ilya Pyatnitskiy, M.D.
University of Texas at Austin
Leander, Texas, United States - HW
Huiliang Wang, Ph.D.
University of Texas at Austin
Austin, Texas, United States
Presenting Author(s)
Main Author(s)
Co-Author(s)
Purpose: Viral vectors, such as adeno-associated vectors (AAV), have become a widely exploited vehicle to express exogenous DNA into target cells due to their high transfection efficiency.1 However, several setbacks of viral vectors severely limited the gene therapy in clinical trials, including immunogenicity, carcinogenesis, limited DNA packaging capacity and high production cost.2 Non-viral vectors have great potential to address many of the limitations of viral vectors, particularly concerning safety, packaging capacity and costs2 but it generally suffers from lower efficiency, especially in primary cells and when utilizing larger-sized plasmids. In particular, efficient cytoplasm diffusion and nucleus delivery of DNA has been the Achilles' heel of non-viral gene delivery.3,4 Specifically, the current non-viral DNA condensation systems are mainly based on electronic interaction between cationic polymer/liposomes and anionic DNA plasmids. However, the electronic interaction is too weak to keep DNA condensed to diffuse through the cytoplasm and transport into the nucleus to initiate gene transcription. The main objective of the current project was to develop a two-stage condensation approach utilizing a reversible, glutathione (GSH)-sensitive crosslinker to form smaller bundled-DNA (b-DNA) that allows for efficient nucleus transport and cytoplasm diffusion for improved gene delivery.
Methods: NPC crosslinkers were synthesized by reaction of 2-hydroxyethyl disulfide and 4-nitrophenyl chloroformate in dichloromethane with the dropwise addition of triethylamine. Purified products were then secondarily reacted with various NH2-PEG-NH2 spacers (PEG1k, PEG2k, PEG3.4K, PEG5k, 4-arm PEG) to form a Glutathione (GSH)-sensitive crosslinker capable of reacting with free amines on plasmid DNA. pCMV-EGFP DNA (6.2 and 11.2 kbp) and crosslinker at a 1:4 w/w ratio were added to PBS pH 8.5 and stirred for 24 hours. Size and zeta potential of b-DNA as a function of reaction time were evaluated and compared to original DNA using DLS (Figure 1). Crosslinking density was evaluated via UV-Vis spectrophotometry by measuring 4-nitrophenol side-product generation over time at λmax 401 nm (Figure 1). In vitro transfection efficiency of b-DNA was evaluated in both HEK293T cells and mice primary neurons. In DIV3 mice primary neurons seeded at 100k cells/well, 1ug of 4-arm, PGE2k and original DNA (6.2 kbp pCMV-EGFP) was complexed with JetOptimus at 1:1 w/v ratio and incubated with cells for 4 hours. After 24 hours cells were fixed and stained for NeuN (red) and DAPI (blue). Fluorescent microscopy was performed and % of GFP+/NeuN+ cells was determined (Figure 2 a and b). In HEK293T cells, 1ug of b-DNA and original DNA (11.2 kbp pCMV-EGFP) was condensed using branched-poly(ethyleneimine) (bPEI) at a N/P ratio of 5 and incubated with cells at 70-80% confluency for 4 hours in FBS-free media. Fluorescence microscopy at 48 hours was performed and % of GFP+ cells was determined by flow cytometry (Figure 2 c and d). Female C57BL/6 mice (n=5) received tail-vein injections of in vivo-Jet PEI nanoparticles formulated with 2 versions of b-DNA (PEG 2K and PEG 5K) and original DNA encoding for firefly luciferase at a dose of 1.5 mg/kg. Live mice were imaged using IVIS at 6, 12 and 24 hours. Luciferase expression in the body was quantified over time. After 24 hours, the organs were extracted for luciferase imaging (Figure 3).
Results: Cross-linking density of various b-DNA increased as a function of reaction time until reaction kinetics plateaued around 24 hours. DNA hydrodynamic size decreased and surface potential increased as cross-linking density of the b-DNA increased over the 24 hour reaction time. Cross-linking DNA using these GSH-sensitive PEG-crosslinkers was capable of producing DNA with sizes less than 100 nm (Figure 1). The polymer/b-DNA complexes exhibited enhanced in vitro gene transfection efficiency compared to polymers/original-DNA complexes. More specifically, in primary neurons the 4-arm PEG and PEG2k b-DNA/polymers complexes showed around 2 times increase in the % of neurons successfully expressing GFP (Figure 2 a and b). Additionally, in HEK293T cells, the b-DNA polyplexes of larger 11.2 kbp plasmid DNA exhibited 2-5-fold increase in GFP expression compared to the original polyplexes (Figure 2 c and d). This in vitro work highlights the improved cytoplasmic diffusion of b-DNA and the increased ability to cross the nuclear membrane in post-mitotic primary cells. Lastly, for in vivo gene delivery both b-DNA formulations (PEG2k and PEG5k) exhibited faster onset and enhanced luciferase expression at 6 hrs. Additionally, use of b-DNA did not alter the biodistribution of the original polyplex and exhibited high levels of expression in the lungs relative to other organs at 24 hours.
Conclusion: This work demonstrated clear improvements in gene delivery efficiency with b-DNA since their smaller size can diffuse through the cytoplasm and into the nucleus more efficiently even in post-mitotic primary neurons. In addition, we have observed a significant improvement in lung-selective gene transfection efficiency by this two-stage condensation approach following intravenous administration. This reversible covalent assembly strategy demonstrates the substantial value of non-viral gene delivery for therapeutic clinical applications.
References: 1. Wang, D.; Tai, P. W. L.; Gao, G. Adeno-Associated Virus Vector as a Platform for Gene Therapy Delivery. Nat. Rev. Drug Discov. 2019, 18 (5), 358–378. 2. Yin, H.; Kanasty, R. L.; Eltoukhy, A. A.; Vegas, A. J.; Dorkin, J. R.; Anderson, D. G. Non-Viral Vectors for Gene-Based Therapy. Nat. Rev. Genet. 2014, 15 (8), 541–555. 3. Lukacs, G. L.; Haggie, P.; Seksek, O.; Lechardeur, D.; Freedman, N.; Verkman, A. S. Size-Dependent DNA Mobility in Cytoplasm and Nucleus. J. Biol. Chem. 2000, 275 (3). 4. Tasset A.; Bellamkonda A.; Wang W.; Pyatnitskiy I.; Ward D.; Peppas N.; Wang H. Overcoming barriers in non-viral gene delivery for neurological applications. Nanoscale. 2022, 14, 3698–719.
Acknowledgements: We thank Dr. Nicholas Peppas, Dr. J. Stachowiak, Dr. L. Suggs, Dr. J. Tunnell, Dr. S. Parekh, Dr. A. Brock and Dr. R. Lu for allowing us to use their equipment and sharing with HEK cells with our lab. Competing Interest Statement: The authors declare that a patent application relating to this work is currently being filed.
Funding: Dr. H. Wang acknowledges funding support from Robert A. Welsh Foundation Grant (No. F-2084-20210327), NIH Mentored Research Scientist Career Development Award (National Institute of Mental Health 1K01MH117490-01), NIH Maximizing Investigators’ Research Award (National Institute of General Medical Sciences 1R35GM147408), Craig Neilsen Foundation and the University of Texas at Austin Startup Fund. Ethics Approval for Animal Use: All procedures involving animals were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals following protocols approved by the University of Texas, Austin Institutional Animal Care and Use Committee.
.jpg) Figure 1: DNA condensation using reversible crosslinkers. (a) The Dynamic Light Scattering (DLS) tests of original DNA and b-DNA; (b) The measurements of DNA crosslinking density and DNA hydration size changes with the reaction time, where crosslinker PEG2000 is used; (c) the measurements of DNA crosslinking density and DNA surface potential changes with the reaction time, where crosslinker PEG2000 is used; (d) The crosslinking density of DNA after reacting with various linkers for 24 h; (e) The final DNA hydration size after reacting with different linkers for 24 h (n=3 per group, one-way ANOVA); (f) The final DNA surface potential after reacting with various linkers for 24 h (n=3 per group, one-way ANOVA). All plots show mean ± SEM unless otherwise mentioned. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant.
Figure 1: DNA condensation using reversible crosslinkers. (a) The Dynamic Light Scattering (DLS) tests of original DNA and b-DNA; (b) The measurements of DNA crosslinking density and DNA hydration size changes with the reaction time, where crosslinker PEG2000 is used; (c) the measurements of DNA crosslinking density and DNA surface potential changes with the reaction time, where crosslinker PEG2000 is used; (d) The crosslinking density of DNA after reacting with various linkers for 24 h; (e) The final DNA hydration size after reacting with different linkers for 24 h (n=3 per group, one-way ANOVA); (f) The final DNA surface potential after reacting with various linkers for 24 h (n=3 per group, one-way ANOVA). All plots show mean ± SEM unless otherwise mentioned. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant.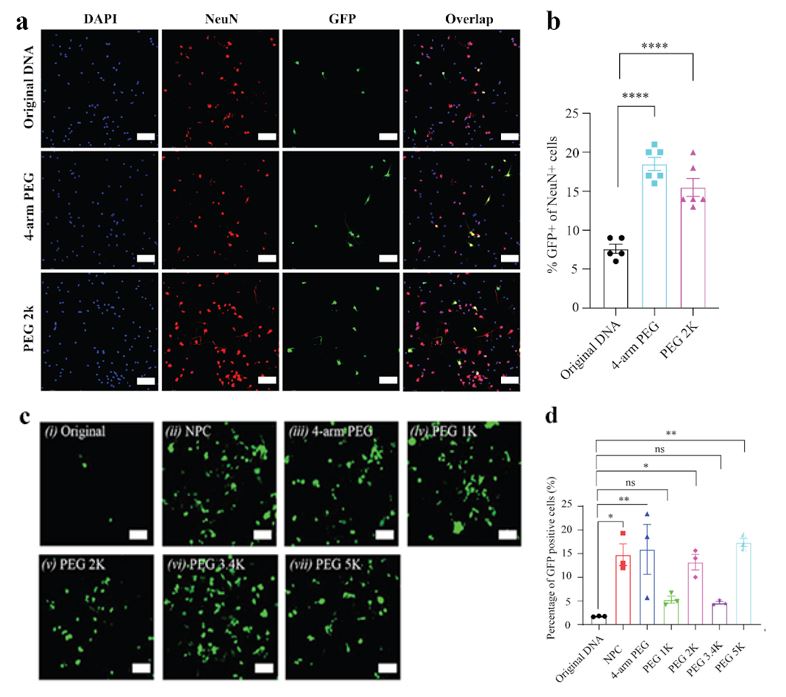 Figure 2: Enhanced green fluorescence protein (GFP) expression of 6.2 kbp b-DNA and 11.2 kbp b-DNA through reversible crosslinkers in vitro. (a) GFP expression images of primary neurons using polymer/DNA(6.2kbp) complexes, where the neuron nucleuses were stained with NeuN (red) and the total cell population nucleuses with DAPI (blue). Scale bar: 100 μm; (b) the corresponding statistics quantification analysis of (a) (n ≥ 2 per group, one-way ANOVA); (c) GFP expression images of HEK cells by various PEI/b-DNA (11.2 kbp) complexes at N/P of 5, (i) original DNA, and condensed DNA via (ii) NPC linker, (iii) 4-arm-PEGylated NPC linker, (iv) PEG1000 NPC linker, (v) PEG2000 NPC linker, (vi) PEG3400 NPC linker, (vii)PEG5000 NPC linker, scale bar: 100 μm, and (d) the corresponding flow cytometry quantification analysis of (c) (n = 3 per group, one-way ANOVA). All plots show mean ± SEM unless otherwise mentioned. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant.
Figure 2: Enhanced green fluorescence protein (GFP) expression of 6.2 kbp b-DNA and 11.2 kbp b-DNA through reversible crosslinkers in vitro. (a) GFP expression images of primary neurons using polymer/DNA(6.2kbp) complexes, where the neuron nucleuses were stained with NeuN (red) and the total cell population nucleuses with DAPI (blue). Scale bar: 100 μm; (b) the corresponding statistics quantification analysis of (a) (n ≥ 2 per group, one-way ANOVA); (c) GFP expression images of HEK cells by various PEI/b-DNA (11.2 kbp) complexes at N/P of 5, (i) original DNA, and condensed DNA via (ii) NPC linker, (iii) 4-arm-PEGylated NPC linker, (iv) PEG1000 NPC linker, (v) PEG2000 NPC linker, (vi) PEG3400 NPC linker, (vii)PEG5000 NPC linker, scale bar: 100 μm, and (d) the corresponding flow cytometry quantification analysis of (c) (n = 3 per group, one-way ANOVA). All plots show mean ± SEM unless otherwise mentioned. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant.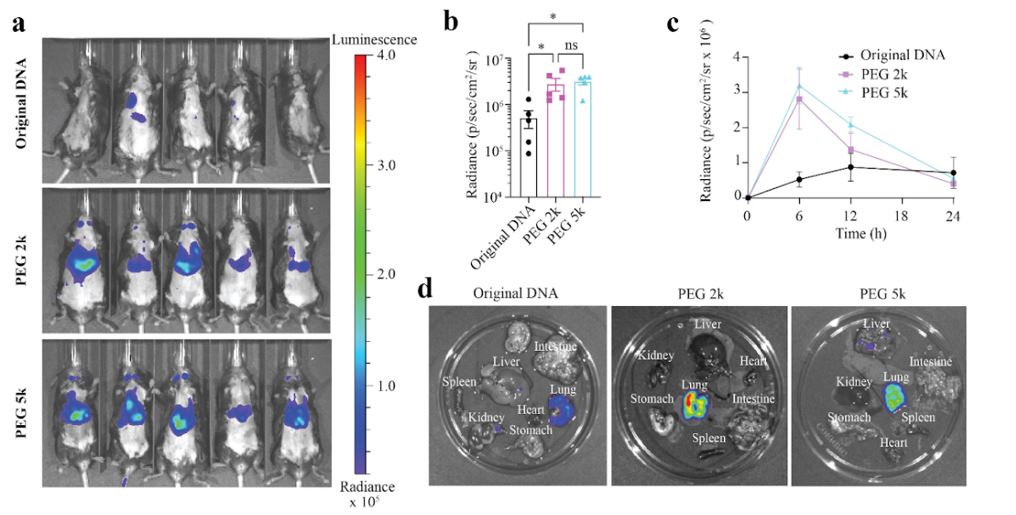 Figure 3: Enhanced in vivo gene delivery efficiency in targeted organs after intravenous administration through RECAST. (a) Bioluminescent images of mice taken with IVIS after 6 h post-injection. (b) Statistical analysis of bioluminescent radiance after 6 h post-injection (n=5, one-way ANOVA, Tukey post-hoc testing). (c) Quantification of luciferase expression in the body over time post-treatment (n=5). (d) Bioluminescent images of extracted organs (heart, liver, spleen, lung, kidney, stomach and intestine) 24h post-treatment, All plots show mean ± SEM. *P < 0.05
Figure 3: Enhanced in vivo gene delivery efficiency in targeted organs after intravenous administration through RECAST. (a) Bioluminescent images of mice taken with IVIS after 6 h post-injection. (b) Statistical analysis of bioluminescent radiance after 6 h post-injection (n=5, one-way ANOVA, Tukey post-hoc testing). (c) Quantification of luciferase expression in the body over time post-treatment (n=5). (d) Bioluminescent images of extracted organs (heart, liver, spleen, lung, kidney, stomach and intestine) 24h post-treatment, All plots show mean ± SEM. *P < 0.05