Discovery and Basic Research
Category: Late Breaking Poster Abstract
(T1430-05-31) Analysis of Pharmacogenetic Polymorphism Effect of ABCB1 2677G >T/A Through Molecular Modeling Approach
Tuesday, October 24, 2023
2:30 PM - 3:30 PM ET
- YL
Yong-Bok Lee
Chonnam National University
Gwangju, Kwangju-jikhalsi, Republic of Korea - JJ
Ji-Hun Jang
Chonnam National University
Gwangju, Kwangju-jikhalsi, Republic of Korea - SJ
Seung-Hyun Jeong
Sunchon National University
Suncheon-si, Cholla-namdo, Republic of Korea
Presenting Author(s)
Main Author(s)
Co-Author(s)
Purpose: P-glycoprotein (P-gp) is widely expressed in the body and is known to be a membrane transporter of multi-drugs with multi-specific recognition properties. Whether or not the pharmacokinetic diversity of drugs that are substrates of P-gp according to the polymorphisms of the ABCB1 gene encoding P-gp has been continuously raised in the past. The aim of this study was to approach the resolution of the pharmacokinetic difference controversy according to the ABCB1 genetic polymorphism (with a focus on the frequently reported missense polymorphism according to 2677G >T/A) through the mechanical function analysis of P-gp using in-silico molecular modeling.
Methods: Based on the human P-gp amino-acid sequence and structural information, P-gp's structures could be modeled. And polymorphic P-gp was created through amino-acid substitution and prepare-protein approach according to base sequence change.
Results: The genetic polymorphism of ABCB1 2677G >T/A (Ala893Ser/Thr) does not correspond to the nucleotide-binding-domain (NBD) or drug-binding-pocket (DBP) of P-gp and does not involve changes in the mechanical conformational of P-gp. Although the amino-acid substitution by c.2677G >T/A caused approximately 30% increased strain in one α-helix hinge between the NBD and DBP of P-gp internal tunnel, no overall structural change was identified (compared to wild-type). This suggested that c.2677G >T/A might have a minor effect on the rate of P-gp inward-to-outward conformational change by increasing the torsional energy, but would not have a significant effect on the general functioning and maintenance of P-gp.
Conclusion: Our findings provide useful insight into resolving the debate about whether ABCB1 genetic polymorphisms are effective covariates and their significance in relation to population pharmacokinetic variability of multiple drugs of P-gp substrates.
References: 1. H. Zhang, H. Xu, C.R. Ashby Jr, et al., Chemical molecular‐based approach to overcome multidrug resistance in cancer by targeting P‐glycoprotein (P‐gp), Med. Res. Rev. 41 (2021) 525–555.
2. S. Dewanjee, T. K. Dua, N. Bhattacharjee, et al., Natural products as alternative choices for P-glycoprotein (P-gp) inhibition, Molecules 22 (2017) 871–963.
3. T. Ishikawa, H. Hirano, Y. Onishi, et al., Functional evaluation of ABCB1 (P-glycoprotein) polymorphisms: high-speed screening and structure-activity relationship analyses, Drug Metab. Pharmacokinet. 19 (2004) 1–14.
4. D. Dickens, A. Owen, A. Alfirevic, et al., ABCB1 single nucleotide polymorphisms (1236C> T, 2677G> T, and 3435C> T) do not affect transport activity of human P-glycoprotein, Pharmacogenet. Genomics 23 (2013) 314–323.
5. Y. Kim, J. Chen, Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation, Science 359 (2018) 915–919.
Acknowledgements:This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2022R1A2C1010245).
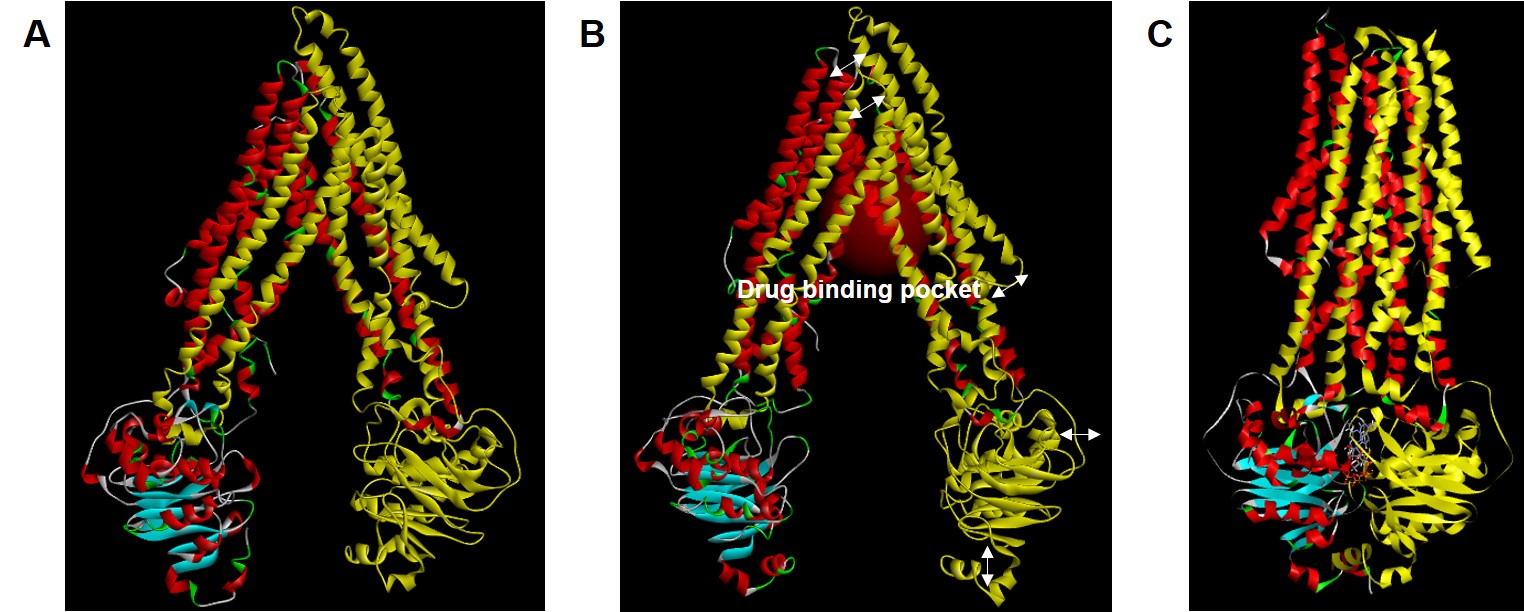 Figure 1. Molecular structures of P-glycoprotein (P-gp) in apo-state (A), ligand-docked state (B), and outward-facing state (C). Arrows indicate major conformational sites in the ligand-docked state compared to the apo-state. For easy comparison of steric changes in each conformational state of P-gp, cassette 1 is labeled in yellow. In the ligand-docked state, red-labeled region represents drug binding pocket.
Figure 1. Molecular structures of P-glycoprotein (P-gp) in apo-state (A), ligand-docked state (B), and outward-facing state (C). Arrows indicate major conformational sites in the ligand-docked state compared to the apo-state. For easy comparison of steric changes in each conformational state of P-gp, cassette 1 is labeled in yellow. In the ligand-docked state, red-labeled region represents drug binding pocket.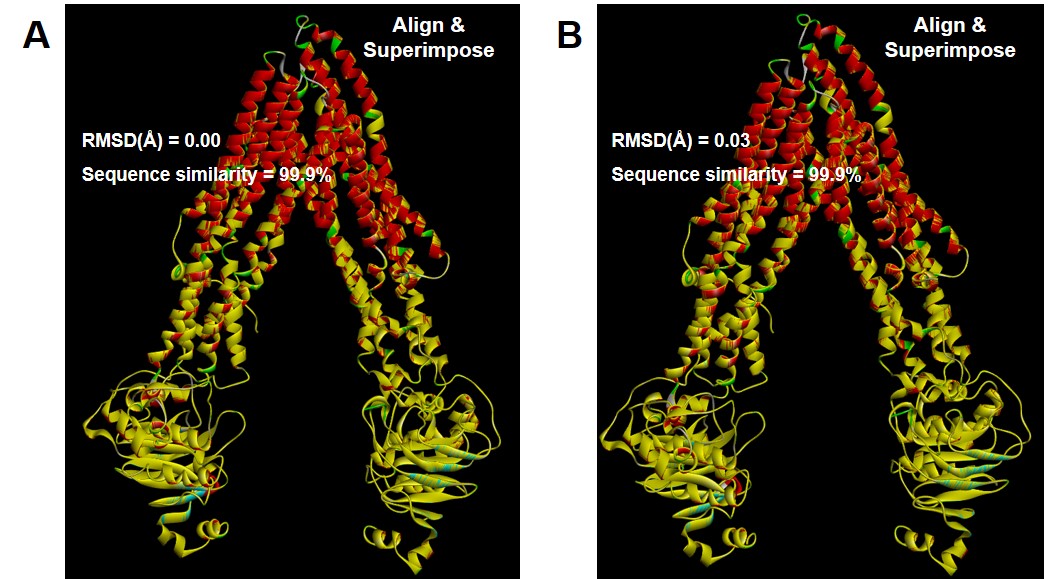 Figure 2. Comparison (alignment and superimpose with wild-type structures) of point polymorphism structures of Ala893Ser (A; ABCB1 2677G>T) and Ala893Thr (B ; ABCB1 2677G>A) in the ligand-docked state of P-glycoprotein. Point polymorphism structures by Ala893Ser/Thr are labeled in yellow.
Figure 2. Comparison (alignment and superimpose with wild-type structures) of point polymorphism structures of Ala893Ser (A; ABCB1 2677G>T) and Ala893Thr (B ; ABCB1 2677G>A) in the ligand-docked state of P-glycoprotein. Point polymorphism structures by Ala893Ser/Thr are labeled in yellow.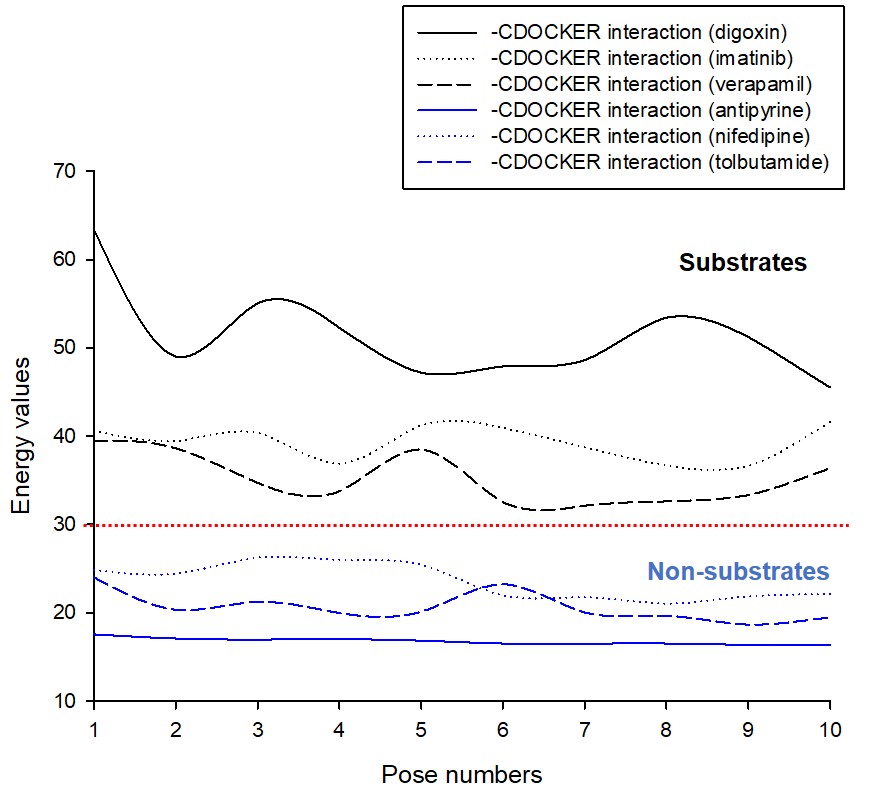 Figure 3. Comparison graph of –CDOCKER interaction energy values according to docking simulation with P-glycoprotein (P-gp) ligand-docked state of drugs corresponding to substrate (digoxin, imatinib, and verapamil) or non-substrate (antipyrine, nifedipine, and tolbutamide) of P-gp. The red dotted line in the graph represents the energy value (30) that approximates the degree of effective substrate interaction with P-gp.
Figure 3. Comparison graph of –CDOCKER interaction energy values according to docking simulation with P-glycoprotein (P-gp) ligand-docked state of drugs corresponding to substrate (digoxin, imatinib, and verapamil) or non-substrate (antipyrine, nifedipine, and tolbutamide) of P-gp. The red dotted line in the graph represents the energy value (30) that approximates the degree of effective substrate interaction with P-gp.