Formulation and Delivery - Biomolecular
Category: Poster Abstract
(W1130-02-11) Interaction between Different Types of Excipients and Protein
- XL
Xun Li, Sr., Ph.D. (he/him/his)
University of Maryland Baltimore
Baltimore, Maryland, United States - XL
Xun Li, Sr., Ph.D. (he/him/his)
University of Maryland Baltimore
Baltimore, Maryland, United States - AO
Asuka A. Orr, Ph.D. (she/her/hers)
University of Maryland
Baltimore, Maryland, United States - AK
Ahmad Karanji, Ph.D. (he/him/his)
University of Maryland
Baltimore, Maryland, United States - FW
Fang Wang, Ph.D. (she/her/hers)
University of Maryland
Baltimore, Maryland, United States - DD
Daniel J. Deredge, Ph.D. (he/him/his)
University of Maryland
Baltimore, Maryland, United States - AM
Alexander D MacKerell, Ph.D.
University of Maryland
Baltimore, Maryland, United States - SH
Stephen Hoag, Ph.D. (he/him/his)
University of Maryland
Baltimore, Maryland, United States
Presenting Author(s)
Main Author(s)
Co-Author(s)
Purpose: There has been a recent surge of interest in the development of high concentration protein formulations (HCPFs), particularly with monoclonal antibodies emerging as a prominent biopharmaceutical subtype in the market. Increasing protein concentration can reduce the required administration volume and enable subcutaneous self-administration, which is a promising product life cycle management strategy that improves patient compliance and offers significant market potential. However, the thermodynamic instability of proteins poses a significant challenge in creating a robust HCPF, resulting in denaturation and aggregation due to self-interaction and crowding effects at high protein concentrations.1-3 This leads to poor biological activity and low bioavailability. To address this issue, excipients such as specific amino acids or sugars are used to influence protein-protein interactions and optimize protein product stability.1 In this study, Chymotrypsin from bovine pancreas and lysozyme from chicken egg white were chosen as model proteins. Either amino acids or sugars were used to stabilize the model proteins with different molecule weights. After performing screening experiments for solubility feasibility, 50 mg/mL was chosen as the highest concentration for both the chymotrypsin and lysozyme formulation. Then, using a rheometer with a temperature ramp1, the thermal stability of the two proteins in the presence and absence of the excipients under investigation was monitored to identify the effect of different excipients.1 Computational modeling using Site Identification by Ligand Competitive Saturation (SILCS)-Biologics4 provide atomistic insights into protein-excipient interactions leading to the stabilization of the proteins in the presence of specific excipients.
Methods: L-Arginine, L-Alanine, Sucrose and Trehalose were weighed and dissolved in 1×PBS buffer separately to make excipient solutions at low (50 mM) and high (500 mM) concentration levels. The pH value was adjusted to 7.4. Chymotrypsin and lysozyme powder were dissolved in the excipient solutions to make protein solutions with a concentration of 50 mg/mL. Proteins in only PBS buffer were used as the control group. Thermal stability was monitored by measuring the change in viscosity through a temperature ramp program. The temperature was ramped up from 10 ℃ to 70 ℃ at a rate of 2 ℃/min, while maintaining a constant shear rate of 50 1/s. The point of a large jump in viscosity value indicates the denaturation temperature point of protein. The data obtained from the viscosity profile was analyzed to compare the thermal stability of protein solutions in different excipient solutions and the control group. Computational modeling: The SILCS-Biologics was applied to chymotrypsin and lysozyme to predict protein-protein interactions (PPI) and excipient binding sites. Briefly, precomputed 3D SILCS FragMaps, representing protein-functional group interaction patterns, are used to map potential excipient binding sites and protein-protein interaction (PPI) probability for the entire protein surface at atomistic level detail. SILCS-Biologics metrics including the distribution of excipient binding sites and their relation to PPI are used to provide atomistic insights into the stabilization of the proteins by the excipients.
Results: A temperature ramp screening program was conducted to identify the denaturation temperature point of the protein resulting from protein-protein interaction that could lead to aggregation. Fig.2A-2B reveals that the jump in viscosity of chymotrypsin shifts to the right in the presence of 500 mM alanine and arginine when compared to the control group. This shift indicates that high concentrations of alanine and arginine have a positive protective effect on chymotrypsin. Fig.2C and Fig.2D show that the jump in viscosity of chymotrypsin in the presence of sucrose and trehalose did not shift when compared to the control group, indicating no significant protective effect on chymotrypsin in PBS buffer. Therefore, it can be concluded that alanine and arginine at 500 mM exhibit a better protective effect for chymotrypsin than sucrose and trehalose. SILCS-Biologics predicted binding sites of alanine and arginine show that the two excipients bind to and cover regions of high PPI propensity whereas these regions are left uncovered by sucrose and trehalose (Fig.1). This suggests that alanine and arginine stabilize chymotrypsin by blocking PPI. As seen in Fig.3 A-D, the jump in viscosity for lysozyme in the presence excipients shift to the right in comparison to the control group, indicating that all four excipients have a stabilizing effect on lysozyme, with higher concentration levels resulting in a better protective effect. In line with the experiments, SILCS-Biologics predicted binding sites of all four excipients show that the excipients bind to and cover regions of high PPI propensity(Fig.1)
Conclusion: In conclusion, the results demonstrate that excipients have varying effects on protein stability, which could be attributed to different protective mechanisms. Molecular modeling of the excipients binding to the proteins using SILCS-Biologics suggest that the excipients stabilize the proteins by binding to regions with high PPI propensity. Future studies should focus on investigating the interaction between protein and excipient, including the binding region, in order to gain a better understanding of their mechanisms and improve their applications in high concentration protein formulation.
References: (1) Jiang, B. Jain, A.; Lu, Hoag, S. W. Probing Thermal Stability of Proteins with Temperature Scanning Viscometer. Molecular pharmaceutics 2019, 16 (8), 3687-3693. DOI: 10.1021/acs.molpharmaceut.9b00598.
(2) Robert G, Strickleya, William J. Lambertb. A review of formulations of commercially available antibodies. Journal of Pharmaceutical Sciences 110, 2021, 2590−2608. DOI: 0.1016/j.xphs.2021.03.017
(3) Tim J. Kamerzell, Reza Esfandiary, Sangeeta B. Joshi, C. Russell Middaugh, David B. Volkin. Protein–excipient interactions: Mechanisms and biophysical characterization applied to protein formulation development. Advanced Drug Delivery Reviews. 63,2011,1118-1159
(4) Jo, S., Xu, A., Curtis, J., Somani, S., and MacKerell, A.D., Jr. “Computational Characterization of Antibody-Excipient Interactions for Rational Excipient Selection using the Site Identification by Ligand Competitive Saturation (SILCS)-Biologics Approach,” Molecular Pharmaceutics, 17: 4323-4333, 2020, 10.1021/acs.molpharmaceut.0c00775, PMC7606568
Acknowledgements: Funding Source: This research was funded by NIH grant 2R44GM130198-02A1. Conflict of Interest: ADM is cofounder and CSO of SilcsBio LLC.
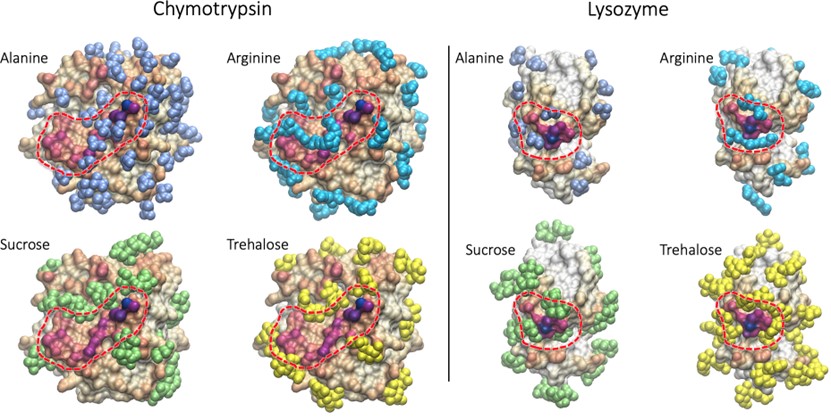 Fig.1 Computationally predicted binding sites of the investigated excipients for chymotrypsin. Alanine, arginine, sucrose, and trehalose excipients are shown in light blue, cyan, green, and yellow VdW representation, respectively. Chymotrypsin (left panel) and lysozyme (right panel) are shown in surface representation with dark purple indicating regions of high PPI propensity, encircled in red dotted lines, and white indicating regions of low or no PPI propensity.
Fig.1 Computationally predicted binding sites of the investigated excipients for chymotrypsin. Alanine, arginine, sucrose, and trehalose excipients are shown in light blue, cyan, green, and yellow VdW representation, respectively. Chymotrypsin (left panel) and lysozyme (right panel) are shown in surface representation with dark purple indicating regions of high PPI propensity, encircled in red dotted lines, and white indicating regions of low or no PPI propensity. 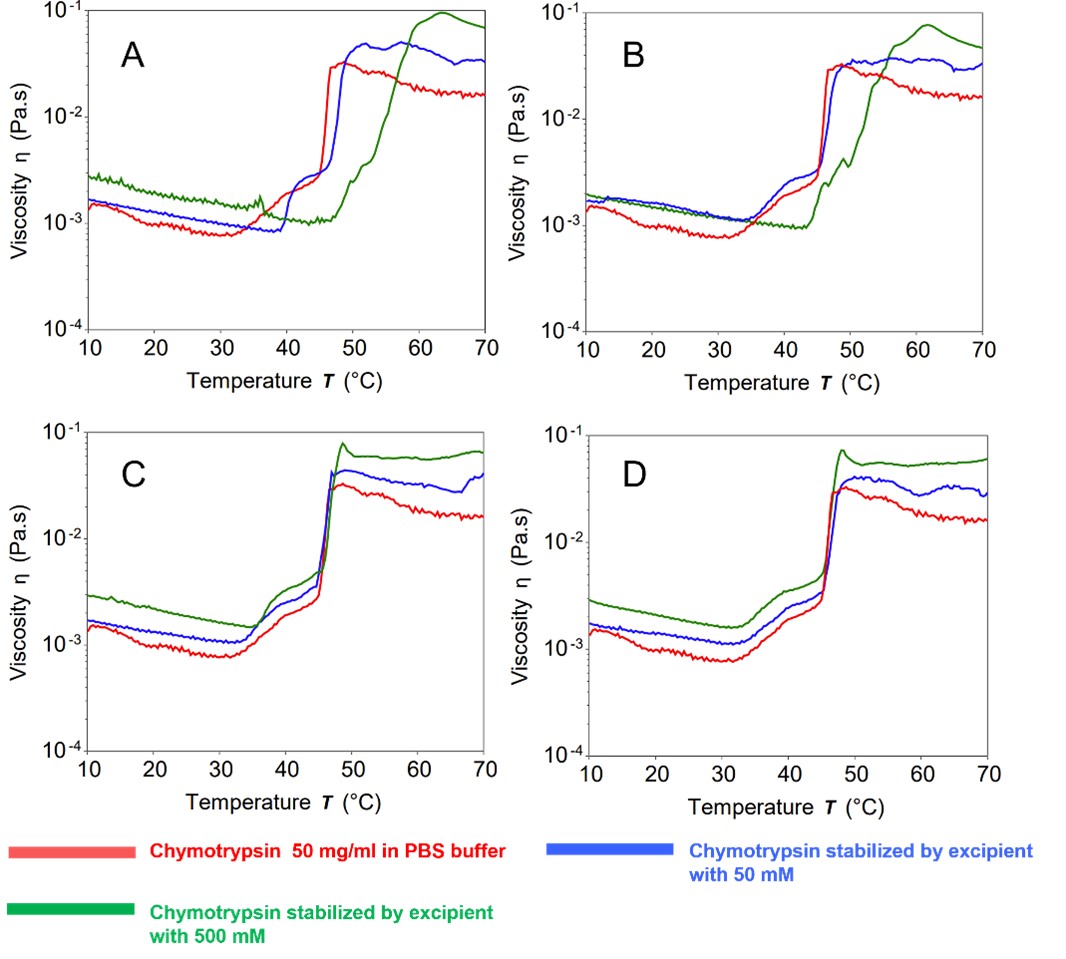 Fig.2 Chymotrypsin as model protein. 2A: Alanine solution, 2B: Arginine solution, 2C: Sucrose solution, 2D: Trehalose solution as stabilizer.
Fig.2 Chymotrypsin as model protein. 2A: Alanine solution, 2B: Arginine solution, 2C: Sucrose solution, 2D: Trehalose solution as stabilizer.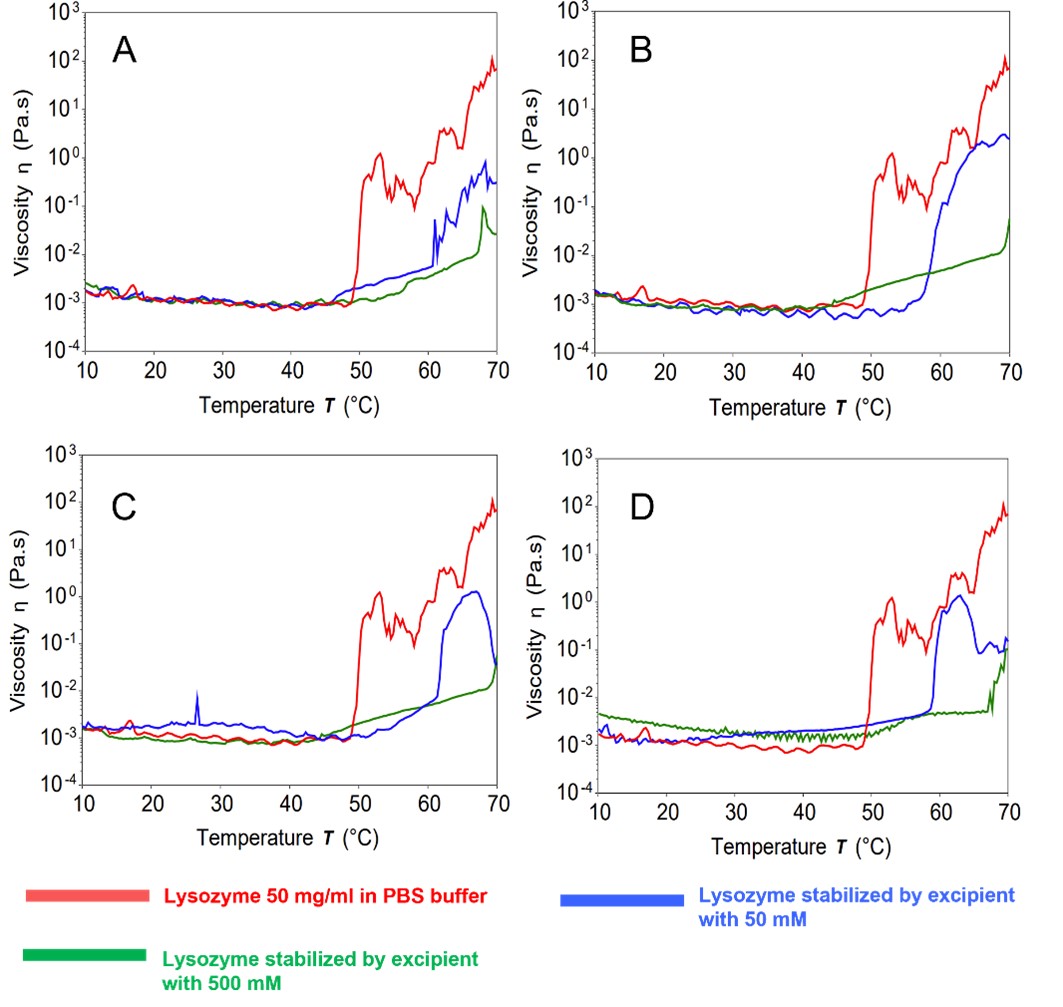 Fig.3 Lysozyme as model protein. 3A: Alanine solution, 3B: Arginine solution, 3C: Sucrose solution, 3D: Trehalose solution as stabilizer.
Fig.3 Lysozyme as model protein. 3A: Alanine solution, 3B: Arginine solution, 3C: Sucrose solution, 3D: Trehalose solution as stabilizer.